SNP Discovery
At present, there are about 80 known SNPs that belong to men in the Little Scottish Cluster. Some of these were discovered accidentally while taking chip based tests. Most however were discovered in tests that attempt to sequence all or part of the DNA of an LSC member. This page will focus on those SNPs discovered via sequencing, but will list the others as well.
1000 Genomes
The 1000 Genomes Project is a large international project to sequence the DNA from individuals of various populations from all over the world. One of those tested, NA12043, belonged to the Little Scottish Cluster, although it was not known at the time.
EthnoAncestry/ScotlandsDNA
EthnoAncestry now ScotlandsDNA was the first company to offer testing for SNPs specific to the LSC. The tests were first offered as individual tests under EthnoAncestry, and then as part of a chip based test by ScotlandsDNA starting in 2012.
The SNPs for these tests were discovered in the DNA of a Wilson Man. Wilson's DNA was sequenced and compared to that of NA12043 to determine which SNPs may be characteristic of the Little Scottish Cluster, and which were unique to the Wilson line in particular. Later testing of Reddin allowed for the general LSC SNPs to be divided into two groups - S190 and equivalents, S424 and equivalents.
| ID | Name | S192 | S424 | S426 | S190 | S308 | S309 | S427 | S421 | S422 | S302 | S303 | S420 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EA1000 | Wilson Sequenced | + | + | + | + | + | + | + | + | + | + | + | + |
| EA2486 | Wilson MRCA 1745 | + | + | + | + | + | + | + | + | + | + | + | - |
| EA1543 | Colson Random LSC | + | + | + | + | + | + | + | - | - | - | - | - |
| EA2998 | Taylor Random LSC | + | + | + | + | + | + | + | - | - | - | - | - |
| EA2997 | Reddin Just beyond LSC | + | + | + | - | - | - | - | - | - | - | - | - |
| - | S192 sample (not LSC) | + | - | - | - | - | - | - | - | - | - | - | - |
Recently, the Chromo2 test at ScotlandsDNA has produced even more results. The Chromo2 test includes more SNPs from Wilson's DNA, and some SNPs were identified that were common only to the Sloan, Chambers and Wilson men in the LSC.
On February 19, 2014 ScotlandsDNA released a spreadsheet (https://www.britainsdna.com/download/C2_2000.zip) with 2000 Chromo2 test results. In the data were the results for 34 S190+ men. The chart below summarizes these results.
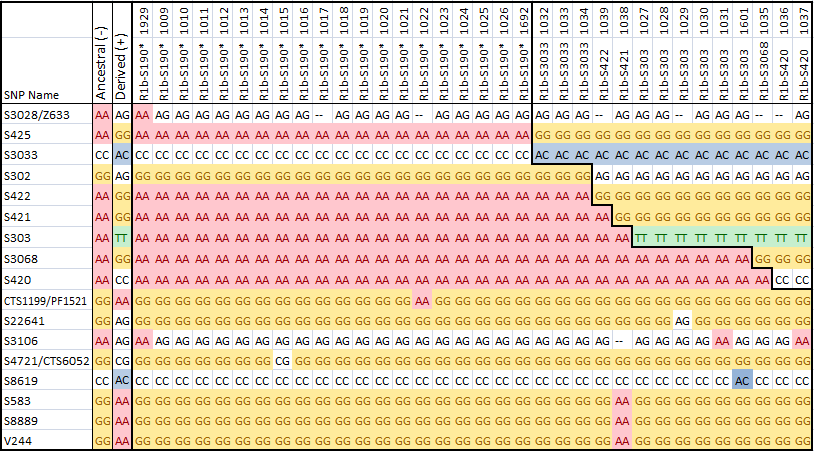
There were 2000 men included in the data released, but only those 34 men who tested S190+ are included in the chart above, and they are numbered in the first row above. There were also over 14000 SNPs in the file, but only those stable SNPs which show any variation within the Little Scottish Cluster are included above.
The SNPs corresponding to the Wilson, Sloan and Chambers group of men are visible in the upper right hand corner of the chart. The Chromo2 test includes a number of SNPs unique to this group, and to the Wilson men in particular. Those columns numbered 1032, 1033 and 1034 must correspond to Sloan and Chambers men. The men following them are all Wilsons, and they fall into a hierarchy of haplogroups.
The data has been released in such as way as it does not identify the men tested. It is possible however to ask ScotlandsDNA if your results were included in the spreadsheet, and what your number is. My own results, Alex Williamson, are in the data as number #1011, while number #1022 corresponds to Steven Colson. If you've been tested at ScotlandsDNA and know your number, please let me know.
The first SNP in the table, S3028/Z633 was thought to be positive for all the men in the Little Scottish Cluster, and negative for everyone else. However, it looks as though we have a S190+ man who is S3028-. This could either be a back mutation, or it could prove that S3028 is downstream of S190, and that this man's line branched off from the main group very early on in the history of the LSC. The identity of this man is confidential, so we'll have to wait until someone else whose identity is known tests in a similar way to find out more.
SNPs S425 through S420 are SNPs which are present in the Sloan, Wilson and Chambers men in the LSC. Sloan and Chambers are both S425+ and S3033+, while the Wilson men are positive for both of these SNPs as well as additional ones.
CTS1199/PF1521 is a SNP which so far has only been found in Steven Colson's results (#1022), while S22641 is unique to the Wilson man (#1029). These SNPs may be present in only these men, but they could also be found in a much larger group of men as well. More testing is needed.
S3106 is an unusual SNP. It is not found outside of the Little Scottish Cluster, but seems to have back-mutated several times within the group.
The remaining SNPs are a little more unstable, and are present in multiple haplogroups outside of the LSC. They may be stable enough however to define clades within the LSC for genealogical purposes. The last 3 SNPs are correlated and seem to mutate together.
FTDNA Walk Through the Y (WTY)
We were lucky to have one member (a Kilgore) take the WTY test at FTDNA back in 2012. This was a time of significant SNP discovery for various R-L21 groups. Unfortunately, no new SNPs were discovered in Kilgore's sample.
FullGenomes.com
In 2013, FullGenomes began to offer Full Y-Chromosome sequencing. Their test offers 50 fold coverage, with a read length of 100. This is sufficient to cover most of the Y-chromosome SNPs, as well as over 400 STRs. As so much of the Y-chromosome is covered, this test offers an amazing opportunity for SNP discovery for both the Little Scottish Cluster as a whole, as well as for private SNPs for a specific lineage. As of November 13, 2013 only one Little Scottish Cluster member, Alex Williamson, is known to have taken the test.For Alex's results, FullGenomes is reporting 292 "private" non-INDEL SNPs. These 292 SNPs come in 4 different levels of quality from 0 to 3 stars (*), with *** being the least reliable. The breakdown of the 292 SNPs is as follows:
| Quality | Likely Genuine? | Count |
| no *s | over 99% | 10 |
| * | over 95% | 13 |
| ** | about 40% | 55 |
| *** | about 10% | 214 |
FullGenomes considers those with 0* or 1* as high quality, and reports that they have found 23 high quality private SNPs in the Y-DNA.
These new SNPs were named FGC3192 to FGC3214.
| SNP | hg19 | anc | der |
| FGC3192 | 3779037 | A | C |
| FGC3193 | 3899172 | G | T |
| FGC3194 | 4511965 | A | G |
| FGC3195 | 4687827 | A | G |
| FGC3196 | 6067820 | A | T |
| FGC3197 | 7952608 | T | C |
| FGC3198 | 7992480 | C | T |
| FGC3199 | 8790228 | A | G |
| FGC3200 | 8980644 | T | A |
| FGC3201 | 9085250 | G | T |
| FGC3202 | 9114725 | G | T |
| FGC3203 | 13128633 | A | C |
| FGC3204 | 13979797 | T | C |
| FGC3205 | 15117619 | G | T |
| FGC3206 | 15452508 | G | A |
| FGC3207 | 17112718 | T | A |
| FGC3208 | 17546935 | C | G |
| FGC3209 | 18017942 | G | A |
| FGC3210 | 21475911 | G | A |
| FGC3211 | 21617233 | A | C |
| FGC3212 | 23275266 | C | G |
| FGC3213 | 26722140 | A | C |
| FGC3214 | 28675156 | T | C |
In terms of INDELs, they found 32 private INDELs of which 2 were high quality.
| SNP | hg19 | anc | der |
| FGC3216 | 14439921 | CAT | C |
| FGC3217 | 21464771 | GTTC | G |
Some of the SNPs and INDELs found were also present in the 1000 genomes person NA12043. These SNPs were identified as follows:
| SNP | hg19 | anc | der |
| FGC3177 | 6826834 | T | C |
| FGC3178 | 3148534 | C | T |
| FGC3179 | 5188687 | C | G |
| FGC3180 | 6408924 | A | G |
| FGC3181 | 6704979 | C | T |
| FGC3182 | 6810313 | A | C |
| FGC3183 | 7323076 | T | C |
| FGC3184 | 7387922 | C | G |
| FGC3185 | 9376352 | G | A |
| FGC3186 | 13378373 | C | T |
| FGC3187 | 14912344 | G | A |
| FGC3188 | 15683936 | T | C |
| FGC3189 | 17361380 | A | C |
| FGC3190 | 21286313 | A | G |
| FGC3191 | 23580097 | G | C |
There was one INDEL in this shared group as well:
| SNP | hg19 | anc | der |
| FGC3215 | 16680097 | GT | G |
The full set of SNP results can also be downloaded, P9UH4A.zip [675 kB].
FTDNA Big Y
This is a brand new test announced on Nov. 9, 2013. It will sequence about 10 million base pairs. Which is less than the FullGenomes test, but so is the cost. So far, as of Nov 29, 2013, we know of 4 LSC members who have ordered the test.
Why Order FullGenomes or the Big Y?
"Why not?" is the far easier question to answer. At $1250 for the FullGenomes test and $500 (going up to $700 in December, 2013) for FTDNA's Big Y, these are not cheap tests! If you can get past their price however, the reason is simple enough, SNP Discovery!
SNPs allow us to unambiguously divide up the the Y-tree with a certainty that is generally impossible with STRs. SNPs are already used to define the haplogroups. Further SNP discovery by members of the LSC, will allow us understand the relationship between the different families. Ultimately, given sufficient testing, they could even be used to divide up the branches on your own family tree!
The diagram below gives a rough overview of current state of SNPs for the LSC.
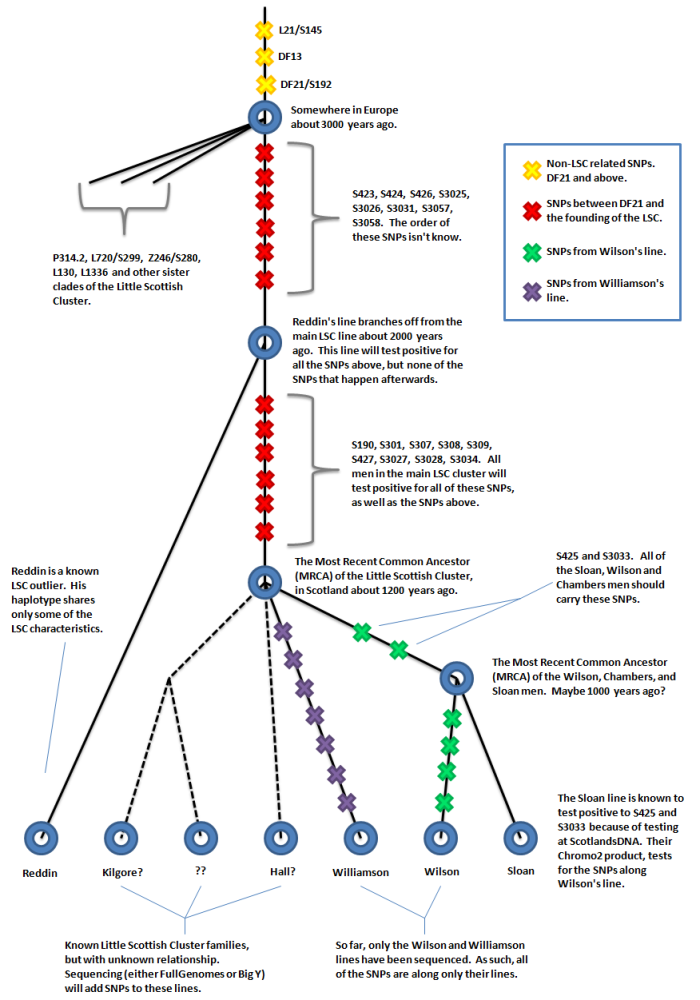
Tests like FullGenomes or the Big Y are necessary to discover new SNPs, but tests like Chromo2 or Geno2.0 can later be used to determine if a man is positive for those SNPs or not.
The FullGenomes test has already generated results. With the Big Y test, we are in uncharted waters. This is a brand new test, for which no one has seen any results. As such, I am somewhat reluctant to encourage anyone to order the test. Having recently had my own Y-DNA sequenced by FullGenomes.com I have some idea of the potential this test has. Both the FTDNA Big Y test, and the FullGenomes test utilize similar technology. If the Big Y can deliver even with the reduced cost, it will be a great test as well.
